PFKFB3 Regulates Oxidative Stress Homeostasis via Its S-Glutathionylation in Cancer.
Seo, M., Lee, Y.H.(2014) J Mol Biology 426: 830-842
- PubMed: 24295899
- DOI: https://doi.org/10.1016/j.jmb.2013.11.021
- Primary Citation of Related Structures:
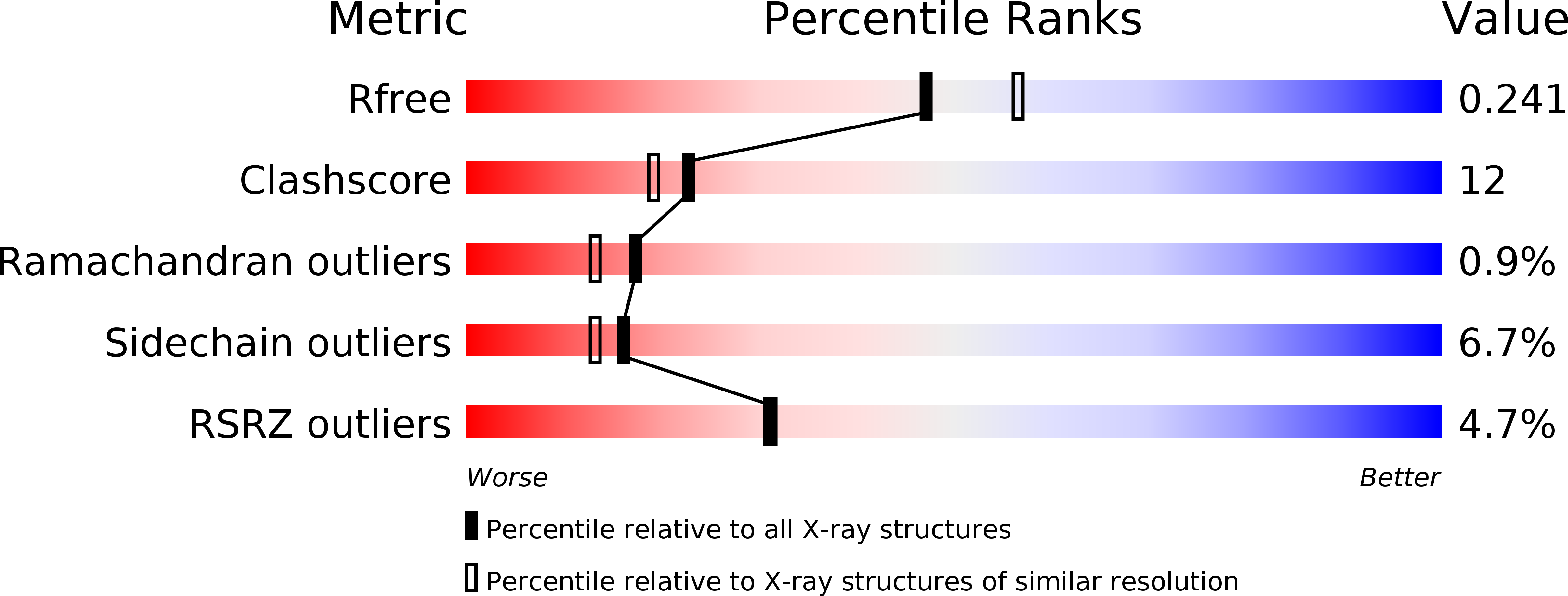
4MA4 - PubMed Abstract:
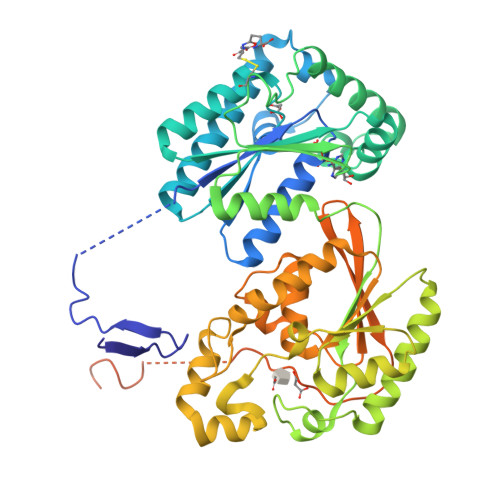
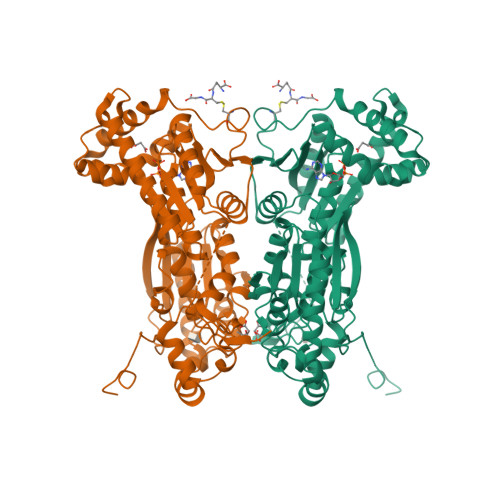
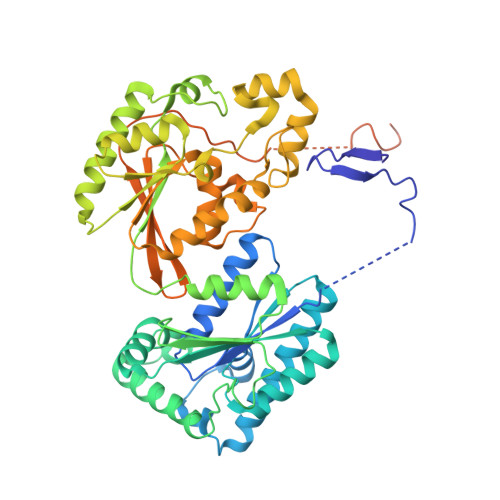
Whereas moderately increased cellular oxidative stress is supportive for cancerous growth of cells, excessive levels of reactive oxygen species (ROS) are detrimental to their growth and survival. We demonstrated that high ROS levels, via increased oxidized glutathione (GSSG), induce isoform-specific S-glutathionylation of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3) at residue Cys206, which is located near the entrance to the 6-phosphofructo-2-kinase catalytic pocket. Upon this ROS-dependent, reversible, covalent modification, a marked decrease in its catalytic ability to synthesize fructose-2,6-bisphosphate (Fru-2,6-P₂), the key glycolysis allosteric activator, was observed. This event was coupled to a decrease in glycolytic flux and an increase in glucose metabolic flux into the pentose phosphate pathway. This shift, in turn, caused an increase in reduced glutathione (GSH) and, ultimately, resulted in ROS detoxification inside HeLa cells. The ability of PFKFB3 to control the Fru-2,6-P₂ levels in an ROS-dependent manner allows the PFKFB3-expressing cancer cells to continue energy metabolism with a reduced risk of excessive oxidative stress and, thereby, to support their cell survival and proliferation. This study provides a new insight into the roles of PFKFB3 as switch that senses and controls redox homeostasis in cancer in addition to its role in cancer glycolysis.
Organizational Affiliation:
Department of Biological Sciences, Louisiana State University, Baton Rouge, LA 70803, USA.